Cellular and molecular mechanisms that support learning and memory in cultured circuits

We want to learn more about the fundamental mechanisms that support learning and memory. We focus on the molecular mechanisms that control synaptic remodeling and the maintenance and modifications of brain circuit connectivity. Synaptic remodeling in the brain during development and in adult life is thought to represent fundamental cellular processes of learning and memory. However, upon abnormal levels of neural activities, it can lead to severe cognitive deficits. The fine line between the mechanisms that produce constructive versus destructive changes in brain circuit connectivity is largely unknown.
To study how synaptic remodeling is regulated, we focus primarily on signaling pathways at excitatory synapses, which are formed on dendritic spines. The methods we use are largely based on neurophotonics, electrophysiology, biochemistry and molecular biology.
We focus on the spatial dynamics and interactions of glutamate receptors (NMDA and AMPA), signaling proteins (CaMKII, netrin) and structural proteins (PSD95, actin, tubulin) that support synaptic plasticity. We combine functional imaging (Ca2+, voltage), optical imaging of fluorescently-tagged proteins, single molecule imaging, fluorescence nanoscopy (STED, PALM), fluorescence lifetime imaging (FLIM) and Forster resonance energy transfer (FRET), patch clamp electrophysiology, biochemistry and molecular biology.
SSeveral genes and proteins that we study are implicated in brain diseases leading to cognitive, psychiatric or neurodegenerative disorders.
This research program exploiting cultured neural circuits is supported by CIHR and a collaborative NeuroNex(NSF) - FRQS program. We collaborate closely with the group of Flavie Lavoie-Cardinal at the CERVO Brain Research Centre.





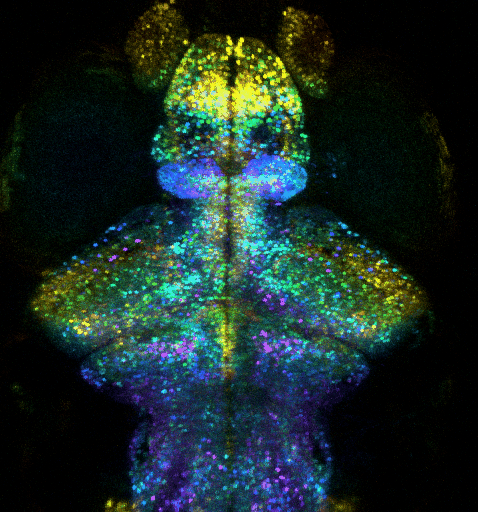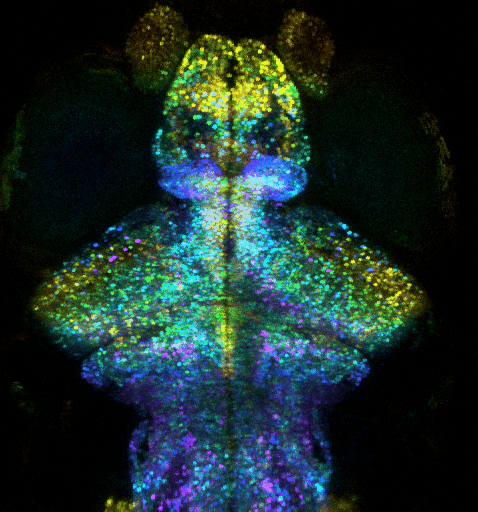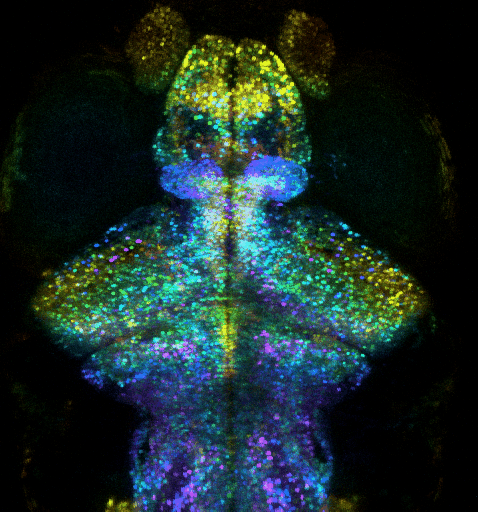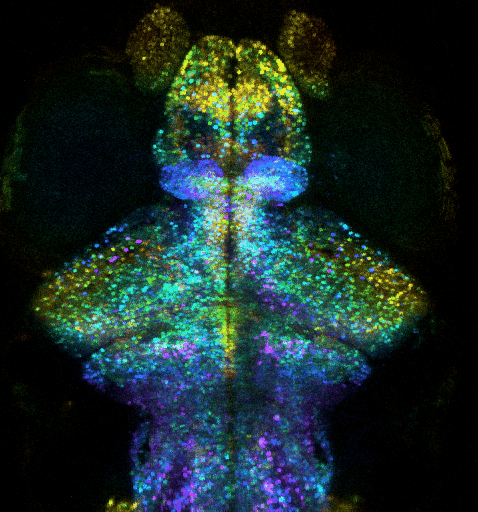
Optogenetic investigation of brain circuit development and plasticity in larval zebrafish

How synaptic and electrical signals enable brain circuits to form, decode sensory inputs, compute information, learn, memorize, and drive behavior are fundamental questions that remain poorly answered. Addressing these problems requires a wide range of models, approaches, and scales.
We recently extended our methods to an animal model providing an “optimal window” to observe synaptic and circuit development, and to contextualize it into brain-wide function, plasticity and behavior: the larval zebrafish (ZF). This model is a strategic choice to investigate how the brain-wide organization and remodeling of neural networks supports learning and memory. We exploit ultra-fast multiphoton microscopy in transparent larval zebrafish expressing fluorescent proteins, such as Ca2+ indicator GCaMP or specifically-targeted fluorescent proteins. We thus combine i) whole-brain functional investigation at neuronal scale and ii) synaptic and sub-synaptic scale high-resolution investigations, while monitoring innate behaviors (tail movement) in response to visual stimuli. We aim to investigate how: i) the brain-wide organization and remodeling of neural networks support learning and memory; ii) synaptic remodeling supports the plasticity of neural networks involved in memory formation, and iii) how the exposome affects these mechanisms. To investigate how the ZF learns and memorizes, we monitor the activity of nearly all neurons in the brain, with two-photon microscopy, in order to exploit an unbiased approach to discover how the memory engram is distributed through the activation and interactions of multiple circuits. In parallel, we zoom into the synaptic scale to quantify any change in number or strength of excitatory and inhibitory synapses, exploiting fish expressing synaptically targeted fluorescent labels. We exploit computational methods to analyze whole-brain and synaptic level datasets.
This combination of strategies enables the investigation of learning at synaptic and brain-wide scales. We also exploit this multiscale approach to examine how exposure to environmental factors impact on brain circuit development. We focus in particular on the gut-brain axis. The zebrafish larva is indeed a strategic model to control the gut microbiota in order to study its communication with the brain.
We are grateful to several scientists who have provided transgenic lines of zebrafish for our experiments, (Pierre Drapeau [U Montreal], Ed Ruthazer [McGill U], Marc Ekker [U Ottawa], Misha Ahrens [Janelia], Tod Thiele [U Toronto], and Gaspard Montandon [Keenan Research Centre for Biomedical Science].
The launch of our Zebrafish Research Program was made possible by the Sentinel North initiative, and is also now supported by NSERC. We collaborate closely with the groups of Patrick Desrosiers, Marie-Ève Paquet and Gabriel Bossé at the CERVO brain research center, and Sylvain Moineau (ULaval) and Pierre Ayotte (CR-CHU).